in-vivo and in-vitro
We employ a holistic approach integrating zebrafish, mice and human pluripotent stem cell based models with unbiased genomics methodologies as well as CRISPR/Cas9-based genome and epigenome editing strategies to understand the regulators of cell-fate decisions.
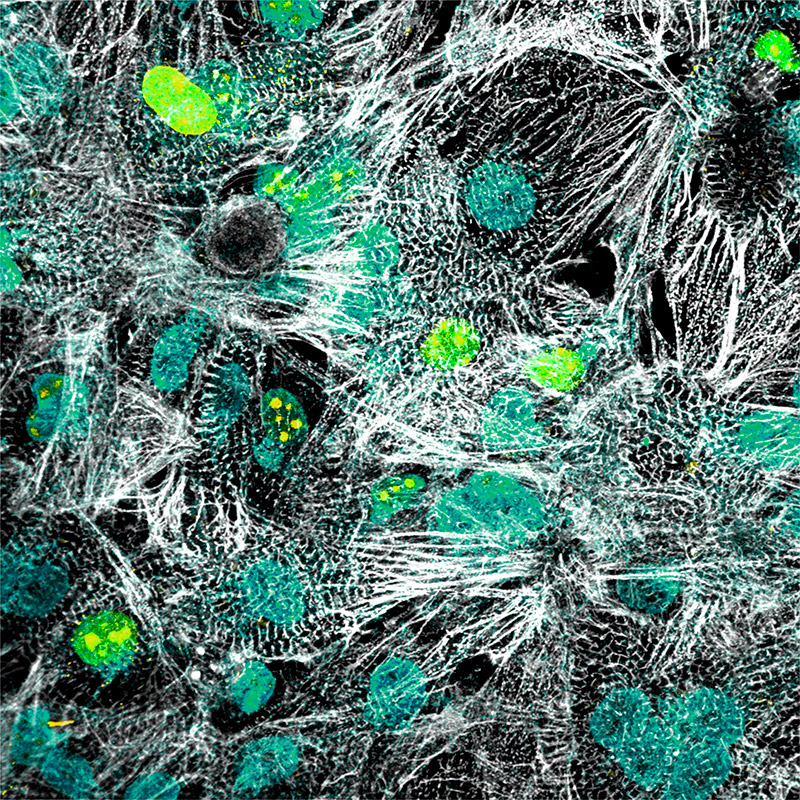
Modeling developmental cell-fate decisions using human pluripotent stem cells
Human pluripotent stem cells are one of the key sources investigated worldwide in the search of the proverbial ‘fountain of youth’ given that every tissue/organ in the body arises from them. By mimicking the signals in a developing embryo in a dish, pluripotent stem cells can be coaxed into activating the genetic networks instructing them to differentiate into functional human cells, which have unlimited therapeutic potential. Realizing this potential, multiple clinical studies are currently under way including cell replacement therapies to improve life-quality in aged individuals. However, our understanding of the genetic networks that control the differentiation processes that lead to the generation of functional human cell types during embryonic development is far from complete. Comprehensive insights on the key genetic determinants of cell-fate decisions and their hierarchy during differentiation is pivotal in understanding embryonic development and to ensure the advancement of safe approaches to generate mature, functional cells for cell-replacement therapies.
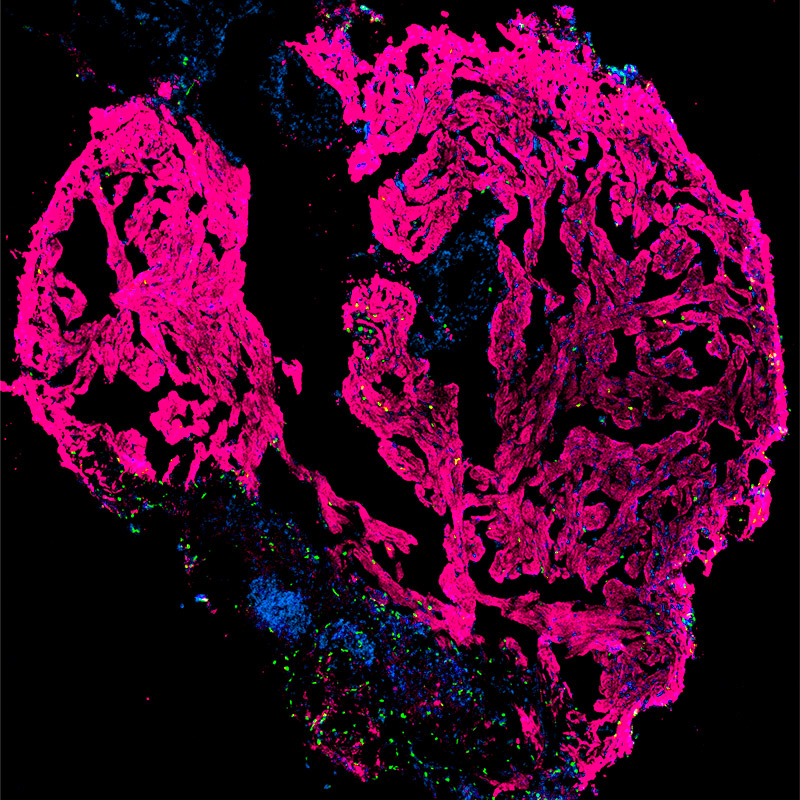
Zerbrafish as an in vivo model of cardiac development and regeneration
In order to study the in vivo function of lncRNAs and other novel genetic elements, we use a collection of cardiovascular reporter zebrafish lines. Importantly, zebrafish can fully regenerate the heart even after a 20% ventricular amputation. However, the molecular network essential to activate and sustain cardiac regenerative response remains poorly elucidated. We aim to understand the role of lncRNAs in cardiac regeneration of the zebrafish heart, and hope to apply these principles to human cardiac regeneration.